2020, Vol. 1, Issue 1, Part A
Phylogenetic and evolutionary revision of some pheasants of northern Pakistan
Author(s): Izaz Ali, Aman Khan, Talmiz Ur Rehman, Minhas Naseer, Shaukat Ali Khan and Humayoun Khan
Abstract: Present study describes the phylogenetic reconstruction and evolutionary relations of seven local pheasant species on the basis of cytochrome b and cytochrome c oxidase I gene sequences. DNA was isolated from blood samples, partial sequences of cytochrome c oxidase (850 bp)and cytochrome b gene (450 bp) were PCR amplified using bird specific universal primers. The nucleotide sequences obtained were compared with that of several members of Phasianidae and other bird species having above 70% homology and phylogenetic trees were constructed using Maximum Likelihood Analysis. The gene sequences obtained from local species have shown multiple conserved regions indicating a common ancestor of these species. The results have suggested a revised systematic position of local pheasants and their phylogenetic distance with related species. The investigated species have been placed in two clades and five sub-clades on the basis of COI gene sequence. The cytochrome b gene sequence has differentiated the species into three clades. Our findings suggest that COI gene can discriminate between similar bird species more efficiently than cytochrome b gene.
Related Graphics: Click here for more related graphics
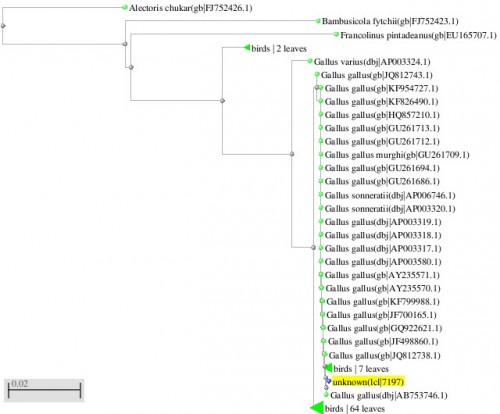
Fig. 1: Phylogenetic tree resulting from the Maximum likelihood analysis of CO1 gene sequence, Parsimony bootstrap 50% majority-rule consensus values were given above the branches and branch length were presented below the branches. Scale bar presented 5 changes per 100 characters. (Sequences of pheasants from Northern Pakistan)
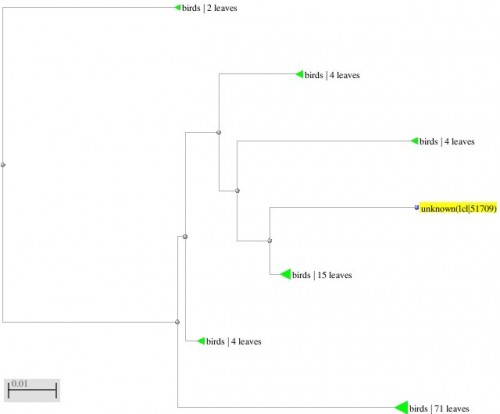
Fig. 2: Phylogenetic tree resulting from the Maximum likelihood analysis of CO1 gene sequence, Parsimony bootstrap 50% majority-rule consensus values were given above the branches and branch length were presented below the branches. Scale bar presented 5 changes per 100 characters. (Sequences of pheasants from Northern Pakistan)
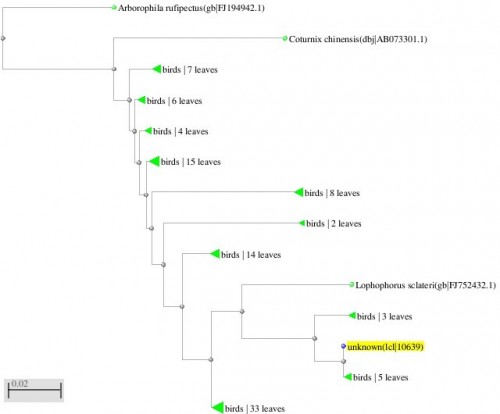
Fig. 3: Phylogenetic tree resulting from the Maximum likelihood analysis of CO1 gene sequence, Parsimony bootstrap 50% majority-rule consensus values were given above the branches and branch length were presented below the branches. Scale bar presented 5 changes per 100 characters. (Sequences of pheasants from Northern Pakistan)
DOI: 10.33545/27080013.2020.v1.i1a.4
Pages: 13-25 | Views: 2177 | Downloads: 1365
Download Full Article: Click Here
How to cite this article:
Izaz Ali, Aman Khan, Talmiz Ur Rehman, Minhas Naseer, Shaukat Ali Khan, Humayoun Khan. Phylogenetic and evolutionary revision of some pheasants of northern Pakistan. Acta Entomol Zool 2020;1(1):13-25. DOI: 10.33545/27080013.2020.v1.i1a.4




